Interview with Prof. Henryk Barthel
When a patient gets a PET scan, usually, the images are taken only after a certain “uptake” waiting period. Can you explain how the radioactive tracer distribution in the body and in the brain change over time from the start of the injection to the end of the standard scan? Why do the patients have to wait?
The radioactive tracer is injected intravenously and travels through the blood flow to reach the brain. The first delivery of the tracer to the brain occurs when it reaches the blood vessels in the brain tissue. The tracer then needs to transfer from the blood vessels into the brain tissue, which can take different amounts of time depending on the specific tracer being used. In order to achieve a steady state where the tracer has evenly distributed throughout the brain, patients typically have to wait for a certain period of time after the injection. The length of the waiting period varies depending on the tracer being used. This waiting “uptake” phase in amyloid PET tracers requires patients to wait 50 – 90 minutes.
Could you explain what dynamic PET scanning in combination with kinetic modeling means?
Dynamic PET scanning in combination with kinetic modeling involves injecting a tracer into the patient directly on the PET scanner and starting the scan immediately. In the brain case, the PET scanner then acquires several so-called dynamic frames of the brain over time, which are used for kinetic modeling. The goal of kinetic modeling is to quantify the PET readout absolutely. For instance, in the case of amyloid imaging, kinetic modeling goal is to provide an absolute number on the concentration of amyloid plaques in the brain. To achieve this, dynamic PET scans are required, as well as the measurement of a time activity curve of the tracer in an artery – the so-called input function.
What measurements, beside a dynamic scan, are needed to perform a full kinetic modeling of the patient?
In addition to a dynamic scan, arterial blood activity samples are usually required for the input curve to perform a full kinetic modeling of the patient.
Could you explain what do kinetic modeling parameters of K1 perfusion and R1 relative delivery mean?
Kinetic modeling can provide various parameters on tracer behavior in the brain, including transfer parameters between different compartments in a compartment model and information on blood flow. K1 and R1 are two such parameters that provide insight into the perfusion and relative delivery of the tracer in the brain, respectively.

If a PET-scan is done in the early phase, right at the start of the injection, what would you see?
In case of amyloid tracers, for instance, early-phase PET images obtained immediately after tracer injection are directly related to the blood flow within the brain. Therefore, often, these images show how different regions of the brain are perfused.
What is cerebral perfusion, regional cerebral blood flow (rCBF) and how does it relate to metabolism in the brain?
This is an important and if not decisive question, because we are in the lucky situation that in the brain blood flow and metabolism / neuronal activity are closely coupled to each other. Cerebral perfusion refers to the blood flow in the brain, which is closely related to the metabolic activity of the neurons. Regional cerebral blood flow (rCBF) is a measure of the blood flow in a specific area of the brain. By measuring rCBF, again, we can estimate the neuronal activity in that area, as blood flow and metabolism are closely coupled. The early phase image therefore allows us to get an image that is similar to FDG tracer (metabolism / neuronal damage) images.
In the context of dementia biomarkers, is there additional value / no additional value in letting one patient be tested for multiple PET-biomarkers, say, one FDG PET neuronal injury biomarker, in addition to an amyloid PET biomarker?
Yes, in many degenerative disorders, both FDG imaging and specific pathology imaging have diagnostic value. A combination of amyloid PET / tau PET and FDG PET, can provide both specific pathology information and neurodegeneration information, which helps to stratify patients. For example, different types of dementia can have the same amyloid pathology but different patterns of neurodegeneration, which can be also used to distinguish these types.
If multiple tracer measurements could be useful, could one perform those in a sequential way, one after another with a certain time in between? What would be problems with this approach?
While it is possible to perform sequential tracer measurements, such an approach would require the patient to undergo multiple imaging sessions, which may be tricky for the patients. Additionally, this would increase radiation exposure and require more staff and more PET system availability.
If multiple tracer measurements could be useful, could one perform those in a dynamic scan with staggered injection approach? What would be problems with this approach?
I don’t think it’s feasible with PET to perform multiple tracer measurements in a dynamic scan with staggered injection approach as I don’t see how a PET scanner would discriminate between different tracers.
If it would be valuable to get the full model tissue parameters (K1, R1, etc.) from a full dynamic scan + auxiliary measurements, is this done in the clinical routine today? Is it done for biotech / pharma studies today to select patients or to perform follow-ups? If not, why not?
The standard practice is to use dynamic scanning with kinetic modeling only in first-in-human clinical testing of novel tracers to fully understand tracer properties. Easier imaging protocols based on static images are then developed for research and clinical routine use. However, there is a debate on whether static images are sufficient to monitor drug effects, and some patients may require dynamic imaging protocols to answer certain questions. Currently, dynamic imaging is not done routinely in clinical practice.
Would you think full dynamic scanning with kinetic modeling would be good in the clinical practice? Would you think it will become clinical practice in the future?
It does look like full dynamic scanning with kinetic modeling is currently not practical for clinical practice due to its demanding nature on the patient, staff, and time. It is unlikely to become a clinical routine approach in the near future.
Can you describe the idea behind the “dual-time-point”, “coffee break” or “dual-time-window” PET imaging method?
The “dual-time-point”, “coffee break” or “dual-time-window” PET imaging method involves acquiring an early scan after tracer injection and a late scan. These two scans can then be used separately as an early perfusion image and a steady state image or be interpolated to obtain an interpolated dynamic scan for analysis using kinetic modeling. These methods have been shown to work well for several tracers, including amyloid tracers and others.
What are the benefits and what are the drawbacks of the dual-time-point method for the clinical routine and trials when compared to full dynamic scanning and the dual tracer method?
Our research shows that the dual-time-point method works well in the clinic and can replace separate FDG scanning for amyloid and tau imaging (Tiepolt et al., 2016; Florek et al., 2018; Tiepolt et al., 2019). We are convinced and our neurologists are also convinced. However, the method is not widely accepted in the community be-yond neuro-PET researchers and needs to be proven with higher level evidence such as multi-center and prospective studies to convince others and authorities for use in wider routine and in clinical trials.
Additionally, there may a biological limitation to the dual-time-point method. For exam-ple, in the healthy brain, blood flow and neuronal activity / metabolism are tightly cou-pled, through the so-called neurovascular coupling. However, this coupling may be dis-turbed in some diseases, such as stroke. That said, in the neurodegenerative diseases that we are studying, such as Alzheimer’s and Parkinsonian syndromes, this coupling holds, and the dual-time-point method can be used. There, for us, it is a valuable tool for clinical diagnosis and research.
Is the dual-time-point method useful in the diagnosis of dementias? What extra-information do you get in this way, that would be difficult to get otherwise?
The prime example is: Imagine a patient is referred to our department for PET scan. A patient with dementia. He or she suffers from cognitive symptoms, which might be severe so that they affect their daily activities. Now the question is, what is the actual underlying disorder?
With the dual-time-point method for the diagnosis of dementias we get extra information that would be difficult to obtain otherwise. For instance, in situations where amyloid load is positive, but the diagnosis is not clear based on amyloid alone, an early after administration blood flow image from the dual time point scanning gives additional information about neurodegeneration, aiding in differential diagnosis. This can help differentiate between different types of dementia, such as Alzheimer’s, Lewy body dementia, and a certain subtype of frontotemporal dementia, which all have different appearances on the blood flow images.
Figure to the right: In a dual time point PET scan, one exploits the dynamic behavior of tracers. In many, e. g. amyloid, tracers a PET image of 10 minutes at from the start of the injection will give an image which is showing the perfusion of the tracers, which is usually coupled to brain metabo-lism. The late scan in contrast shows the usual clinical routine image. Adapted from: V. Garibotto, “FDG and Amyloid PET in Dementia”, 2021 SGNM series Swiss Hybrid Imaging
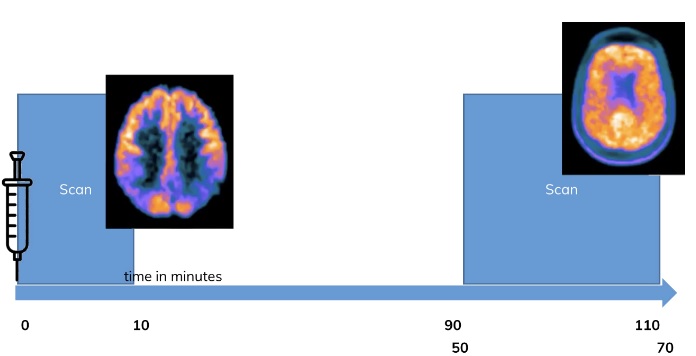
Do you use the dual-time-point method in your clinical routine? Do you use the method in clinical studies? Do you know of other centers that use the method?
The dual-time-point method is standard routine practice in our clinic where it is used for amyloid and tau imaging. It is not yet used in all the clinical trials due to the previously discussed need for more prospective multi-center studies to establish its effectiveness there. I know of several other centers who routinely use this method al-ready, and I believe it is part of the data collection in the ADNI/AIBL study. We have established it in our center and the method is found to be helpful and demanded by our neurologists for better diagnostic accuracy. They want to see these images and they also demand that we put it in our reports. Overall, the method is already established in several centers such as ours here in Leipzig and gaining acceptance in other centers.
If you had a compact brain PET-only system like NeuroLF, could you imagine performing the dual-time-point method with such a PET system placed directly in the tracer administration room? How would this impact the clinical workflow, compared to how you would have to do it today? Would there be benefits / drawbacks?
Using a compact brain PET-only system like NeuroLF in the tracer administration room would have a positive impact on the clinical workflow in dual-time-point scanning. It would eliminate the need for patients to be transferred to the scanner room twice, making the procedure more efficient and convenient for staff and patients. This would also free up time for other patients to be scanned, as the interleave scheduling of patients would no longer be necessary, which can get tricky. Overall, again, a dedicated scanner in or close to the injection room would be a great improvement for this clinical workflow.
Some authors say (Joseph-Mathurin et al., 2018; Bilgel et al., 2020), that, when trying to extract the maximum information out of one tracer injection, the kinetic modelling parameters (in particular R1) are better than the early phase images from a dual-time-point PET measurement as indicator of perfusion and as biomarker for neuronal injury. Would you agree? Would those be better suited for clinical practice or clinical studies, than a single early phase dual-time-point PET image?
In the literature, following authors (among others) have also tested the dual-time-point method on a variety of tracers, including claimed successes with:
Amyloid tracers such as [11C]Pittsburgh compound B (Joseph-Mathurin et al., 2018; Fu et al., 2014; Rostomian et al., 2011), [18F]
Florbetaben (Bullich et al., 2018; Daerr et al., 2017; Heeman et al., 2019; Tiepolt et al., 2016), [18F], Florbetapir (Albano et al., 2021), [18F], Flutemetamol (An et al., 2021), [18F], Florapronal (Jeong et al., 2019)
Tumor hypoxia tracer [18F] FMISO (Grkovski et al., 2017)
Tau tracers [18F]THK5317 (Rodriguez-Vieitez et al., 2017), [18F]THK5351 (Brendel et al., 2017), [18F]PI-2620 (Beyer et al., 2020)
Astrocyte tracer [11C]DED (Rodriguez-Vieitez et al., 2016), [18F]THK5351 (Brendel et al., 2017), [18F]PI-2620 (Beyer et al., 2020)
Parkinsonism tracer [18F]FP-CIT (An et al., 2021)
The method seems quite generic for multiple tracers – Would you agree? Why is that? What parameter is important in a tracer, so that it works?
Would you see other clinical application for the dual-time-point method, besides the mentioned application in diagnosis of Alzheimer’s disease?
Several authors have published the desire (Peretti et al., 2019 (a); Peretti et al., 2019 (b); Bilgel et al., 2020) to establish more quantification with full dynamic scanning and kinetic modeling in the clinical practice. In dual-time-point / window protocols, one acquires with a time gap that misses data, when compared to a full dynamic scan. In theory, from the two dual-time-point measurements (early phase and later regular steady state phase), would it be possible to interpolate the missing data in-between, to allow for a non-invasive and simplified, yet full quantification?
Would you recommend the dual-time-point protocol to other large centers of neuro-nuclear medicine like yours? Would you recommend it to smaller scale centers of nuclear medicine?
Prof. Henryk Barthels
Prof. Henryk Barthel is a nuclear medicine physician and a researcher in the field of neuro-nuclear imaging. He is a senior physician of the Department of Nuclear Medicine at the University Hospital of Leipzig, Germany (Director and Chairman: Prof. Dr. med. Osama Sabri). He obtained his medical degree from the University of Leipzig in 1996, and he has been working in the field of multi-modal neuro-nuclear imaging since then.
His research focuses on the use of PET/CT and PET/MR imaging for the diagnosis and management of neurodegenerative diseases. He has authored and co-authored numerous scientific publications in the field of neuro-nuclear imaging, and he is an active member of several national and international scientific organizations in the field.
